How To Change Na To Na In R
Missing values in data science ascend when an observation is missing in a cavalcade of a information frame or contains a character value instead of numeric value. Missing values must be dropped or replaced in order to depict correct conclusion from the data.
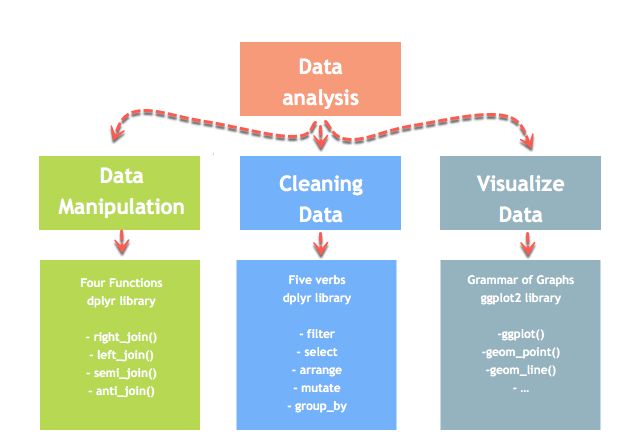
In this tutorial, we will acquire how to deal with missing values with the dplyr library. dplyr library is role of an ecosystem to realize a data analysis.
In this tutorial, you volition acquire
- mutate()
- Exclude Missing Values (NA)
- Impute Missing Values (NA) with the Mean and Median
mutate()
The fourth verb in the dplyr library is helpful to create new variable or alter the values of an existing variable.
We will keep in two parts. We volition learn how to:
- exclude missing values from a information frame
- impute missing values with the hateful and median
The verb mutate() is very piece of cake to use. We can create a new variable post-obit this syntax:
mutate(df, name_variable_1 = condition, ...) arguments: -df: Data frame used to create a new variable -name_variable_1: Name and the formula to create the new variable -...: No limit constraint. Possibility to create more than one variable inside mutate()
Exclude Missing Values (NA)
The na.omit() method from the dplyr library is a simple mode to exclude missing observation. Dropping all the NA from the data is easy simply it does not hateful it is the almost elegant solution. During assay, it is wise to use variety of methods to deal with missing values
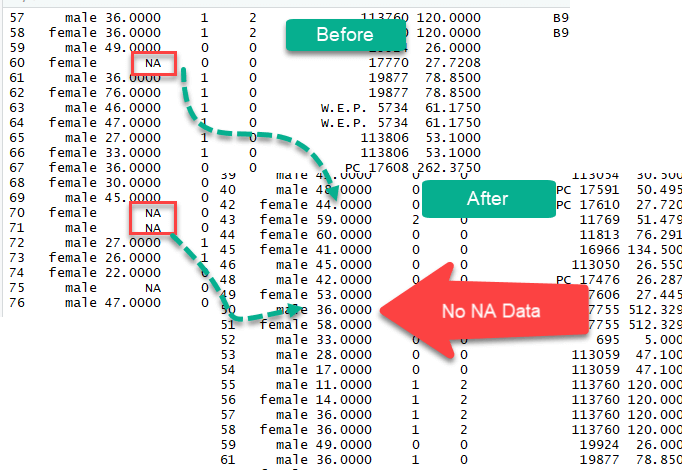
To tackle the problem of missing observations, we will employ the titanic dataset. In this dataset, we have access to the data of the passengers on lath during the tragedy. This dataset has many NA that demand to exist taken care of.
We will upload the csv file from the cyberspace and then cheque which columns have NA. To return the columns with missing data, nosotros tin can use the post-obit code:
Permit'due south upload the information and verify the missing data.
PATH <- "https://raw.githubusercontent.com/guru99-edu/R-Programming/master/test.csv" df_titanic <- read.csv(PATH, sep = ",") # Return the column names containing missing observations list_na <- colnames(df_titanic)[ apply(df_titanic, 2, anyNA) ] list_na
Output:
## [1] "age" "fare"
Here,
colnames(df_titanic)[apply(df_titanic, 2, anyNA)]
Gives the proper name of columns that exercise non have information.
The columns age and fare accept missing values.
We tin drop them with the na.omit().
library(dplyr) # Exclude the missing observations df_titanic_drop <-df_titanic %>% na.omit() dim(df_titanic_drop)
Output:
## [1] 1045 xiii
The new dataset contains 1045 rows compared to 1309 with the original dataset.
Impute Missing data with the Mean and Median
We could as well impute(populate) missing values with the median or the hateful. A expert practise is to create two separate variables for the mean and the median. Once created, we can replace the missing values with the newly formed variables.
We will use the apply method to compute the hateful of the column with NA. Allow's see an instance
Footstep 1) Before in the tutorial, we stored the columns name with the missing values in the list called list_na. We will utilise this list
Step 2) Now we need to compute of the mean with the argument na.rm = TRUE. This argument is compulsory because the columns accept missing information, and this tells R to ignore them.
# Create mean average_missing <- employ(df_titanic[,colnames(df_titanic) %in% list_na], two, mean, na.rm = TRUE) average_missing
Lawmaking Explanation:
We pass iv arguments in the utilize method.
- df: df_titanic[,colnames(df_titanic) %in% list_na]. This code volition return the columns proper noun from the list_na object (i.e. "age" and "fare")
- ii: Compute the function on the columns
- mean: Compute the mean
- na.rm = Truthful: Ignore the missing values
Output:
## age fare ## 29.88113 33.29548
We successfully created the mean of the columns containing missing observations. These two values will be used to replace the missing observations.
Step 3) Replace the NA Values
The verb mutate from the dplyr library is useful in creating a new variable. We don't necessarily want to alter the original cavalcade so nosotros can create a new variable without the NA. mutate is easy to use, nosotros only choose a variable name and define how to create this variable. Here is the complete code
# Create a new variable with the mean and median df_titanic_replace <- df_titanic %>% mutate(replace_mean_age = ifelse(is.na(age), average_missing[1], age), replace_mean_fare = ifelse(is.na(fare), average_missing[2], fare))
Code Explanation:
We create two variables, replace_mean_age and replace_mean_fare equally follow:
- replace_mean_age = ifelse(is.na(historic period), average_missing[1], historic period)
- replace_mean_fare = ifelse(is.na(fare), average_missing[2],fare)
If the column historic period has missing values, then replace with the showtime chemical element of average_missing (mean of age), else proceed the original values. Aforementioned logic for fare
sum(is.na(df_titanic_replace$age))
Output:
## [1] 263
Perform the replacement
sum(is.na(df_titanic_replace$replace_mean_age))
Output:
## [one] 0
The original column age has 263 missing values while the newly created variable take replaced them with the mean of the variable historic period.
Step 4) Nosotros can supercede the missing observations with the median as well.
median_missing <- apply(df_titanic[,colnames(df_titanic) %in% list_na], two, median, na.rm = Truthful) df_titanic_replace <- df_titanic %>% mutate(replace_median_age = ifelse(is.na(age), median_missing[one], age), replace_median_fare = ifelse(is.na(fare), median_missing[ii], fare)) caput(df_titanic_replace)
Output:

Pace 5) A big information prepare could accept lots of missing values and the higher up method could be cumbersome. Nosotros can execute all the in a higher place steps above in one line of code using sapply() method. Though nosotros would not know the vales of mean and median.
sapply does not create a data frame, then we can wrap the sapply() function within information.frame() to create a data frame object.
# Quick code to supplant missing values with the mean df_titanic_impute_mean < -data.frame( sapply( df_titanic, role(x) ifelse(is.na(x), mean(x, na.rm = True), x)))
Summary
We have 3 methods to deal with missing values:
- Exclude all of the missing observations
- Impute with the mean
- Impute with the median
The following tabular array summarizes how to remove all the missing observations
| Library | Objective | Lawmaking |
|---|---|---|
| base | List missing observations | colnames(df)[utilise(df, two, anyNA)] |
| dplyr | Remove all missing values | na.omit(df) |
Imputation with mean or median tin be done in 2 ways
- Using utilize
- Using sapply
| Method | Details | Advantages | Disadvantages |
|---|---|---|---|
| Step by step with apply | Check columns with missing, compute mean/median, shop the value, replace with mutate() | You know the value of ways/median | More execution time. Can exist slow with large dataset |
| Quick manner with sapply | Apply sapply() and data.frame() to automatically search and replace missing values with mean/median | Brusk lawmaking and fast | Don't know the imputation values |
Source: https://www.guru99.com/r-replace-missing-values.html
Posted by: bynumraimad.blogspot.com

0 Response to "How To Change Na To Na In R"
Post a Comment